|
TINKER:
A complete package for molecular mechanics, dynamics and modeling of
molecules, especially biomacromolecules. Tinker has the ability to use
any of several common parameter sets, such as Amber (ff94, ff96, ff98,
ff99), CHARMM (19, 22 and 22-CMAP), Allinger MM (MM2-1991 and MM3-2000),
OPLS (OPLS-UA, OPLS-AA and OPLS-AA/L), as well as our AMOEBA polarizable
atomic multipole force field. Tinker implements a variety of novel
algorithms including distance geometry with fast metrization and
Gaussian trial distances, Elber's reaction path method, global
optimization via our Potential Smoothing and Search algorithms,
molecular dynamics with simulated annealing and stochastic dynamics
options, particle mesh Ewald summation, Monte Carlo minimization,
atomic multipole treatment of electrostatics with explicit dipole
polarizability, Eisenberg-McLachlan ASP and GB/SA continuum solvation
models, and truncated Newton TNCG local energy minimization.
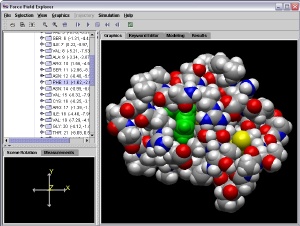
FORCE FIELD EXPLORER:
A Java-based GUI for Tinker, which includes molecular visualization
as well as the ability to launch Tinker calculations from a graphical
interface. FFE uses a socket mechanism for communication between the
GUI and Tinker programs, allowing local monitoring of jobs running on
remote machines. The program can also download organics and drugs from
PubChem and the NCI, and biopolymer structures from the PDB.
SLEUTH:
A small set of programs that allow creation of PDB file summaries and
searching of a dataset extracted from the PDB for structural motifs. For
example, SLEUTH can find all examples of type I turns that match some
generalized sequence pattern. An additional program implements the
Eisenberg 3D profile algorithm. The /data subdirectory contains a set of
summary files with dihedral angle values, secondary structure and solvent
exposure data by residue for many of the current PDB structures.
PROPAK:
A program to attack the "inverse" protein folding problem
via modeling of the side chain packing in proteins. Allows the user to
redesign a portion of a known structure using a library of side chain
rotamers. Provides a list of the amino acid sequences that are predicted
be compatable with an input tertiary fold. The PROPAK algorithm was one
of the first attempts to attack this general protein design problem.
The original program, although still useful, has not been under active
development for some time and is largely superceded by methods from
the labs of Steve Mayo, David Baker, and many others.
QSAR:
A program to derive QSAR models via regression analysis. The analysis
can be performed by ordinary least squares (OLS), principal components
(PCR) or partial least squares (PLS). "Leave-1-out"
cross-validation is performed for all three types of analysis. For OLS
and PCR, variable selection can be performed via simulated annealing on
predictive power. In all cases, the final QSAR model can be used to make
predictions for a test set distinct from the training set. Several training
set data tables, including Garland Marshall's VALIDATE set for prediction
of binding in macromolecular complexes, are provided as examples.
|
|
|
|
|
| |
Ryan Miller
Graduate Student, Computer Science
University of California, Irvine
ryan.miller@wustl.edu
|
Chris Ho
Senior Research Scientist
Qubit Pharmaceuticals
cho@newdrugdesign.com
|
|
|
Yuanjun "Sherlock" Shi
Graduate Student, Chemistry
Yale University
yuanjun@wustl.edu
|
Borna Novak
MSTP Student
Washington University School of Medicine
bnovak@wustl.edu
|
|
|
Marie Laury
Bioinformatics Scientist, GTAC
Washington University School of Medicine
majkutml@gmail.com
|
Josh Rackers
Machine Learning Scientist
Prescient Design
joshrackers@gmail.com
|
|
|
Aaron Gordon
Software Engineer
Boeing, Defense, Space & Security, St. Louis
aarongordon@wustl.edu
|
Chao Lu
Lead, Force Field Development
ByteDance
chaolupersonal@gmail.com
|
|
|
Kailong Mao
Associate Software Engineer
Allscripts Healthcare Solutions, Raleigh, NC
kailong.mao@gmail.com
|
Brendan McMorrow
Staff Software Engineer
DeepMind Technologies Limited, London
mcmobc2@gmail.com
|
|
|
|
Brooke Husic
Fellow, Center for Theoretical Science
Princeton University
brookehusic@gmail.com
|
Pooja Suresh
Biophysicist & Data Scientist, CytoTronics
Boston, Massachusetts
pooja.suresh@ucsf.edu
|
|
|
Laura Watkins
Data Scientist, Kemper
Chicago, Illinois
lcwatkins21@gmail.com
|
Justin Xiang
Chief Investment Officer
Syno Capital, New York, NY
justin.xiang@synocapital.com
|
|
|
Chris Mejias
Resident Physician, Diagnostic Radiology
Barnes-Jewish Hospital, Saint Louis, MO
mejiasc@wusm.wustl.edu
|
Tyler Ponder
Software Engineer, AWS S3 Storage
Amazon Web Services, New York, NY
tponder11@gmail.com
|
|
|
Jeffrey Bigg
Software Engineer
Propel, Inc., Brooklyn, NY
jbigg.2012@gmail.com
|
Victoria Wang
Clinical Fellow, Brigham & Women's Hospital
Boston, Massachusetts
victoria.wang.912@gmail.com
|
|
|
Yong Huang
ETL and Data Conversion Engineer
Three Rivers Systems, Inc.
yhuang01@gmail.com
|
Dave Gohara
Technical Staff, Fungible, Inc.
Santa Clara, California
dgohara@slu.edu
|
|
|
Chuanjie Wu
Research Leader
Schrodinger, New York, NY
chuanjiewu@gmail.com
|
Sergio Urahata
Institute of Physics
University of Sao Paulo, Brazil
surahata@gmail.com
|
|
|
Mike Schnieders
Associate Professor, Biomedical Engineering
University of Iowa
michael-schnieders@uiowa.edu
|
Pengyu Ren
Professor, Biomedical Engineering
University of Texas, Austin
pren@mail.utexas.edu
|
|
|
Alan Grossfield
Associate Professor, Biochemistry & Biophysics
University of Rochester Medical School
alan_grossfield@urmc.rochester.edu
|
Peter Bagossi
Associate Professor, Biochemistry & Molecular Biology
University of Debrecen, Hungary
bagossi@med.unideb.hu
|
|
|
Enoch Huang
Vice President, Machine Learning & Computational Sciences
Pfizer R&D, Cambridge, MA
enoch_huang@cambridge.pfizer.com
|
Reece Hart
Chief Technology Officer, MyOme
Palo Alto, California
reecehart@gmail.com
|
|
|
Rohit Pappu
Professor, Biomedical Engineering
Washington University, St. Louis
pappu@wustl.edu
|
Yong Kong
Associate Director, Bioinformatics
W. M. Keck Foundation Laboratory, Yale University
yong_kong@yale.edu
|
|
|
Megan Fedders
Director, Analytics & Value Management
Evernorth Direct Health, Hartland, WI
meganfedders@yahoo.com
|
Mike Dudek
Founder & Chief Scientist
Hexadecapole, San Diego, CA
mdudek@hexadecapole.com
|
|
|
Margaret Goodman
Professor, Biology
Wittenberg University
mgoodman@wittenberg.edu
|
Uma Kuchibhotla
Chief Scientific Officer, Biotechnology
Eli Lilly and Company
kuchibhotla_uma@lilly.com
|